Cytomine ULiège Research & Development
@ Montefiore Institute (Dept. EE & CS), University of Liège, Belgium
At the Montefiore Institute (University of Liège), we are software developers and computer science researchers developing machine/deep learning algorithms and big data software modules to make life easier for multidisciplinary teams who have to deal with very large imaging data. Our software tools enable remote collaboration through sharing of images, algorithms, and quantitative results over the web. Our developments are used worldwide in biomedical research and beyond.
Our team continuously contributes to the Cytomine open-source project initiated by our research unit in 2010. We maintain the free, open-source (Apache 2.0) Cytomine software and we develop specific modules and experimental features driven by our international research projects and collaborations. In addition, we develop experimental features driven by our research projects and collaborations.
Ongoing research & results
The Cytomine open-source software (with a permissive licence) is a rich internet application using modern web and container technologies, geospatial and noSQL databases, and machine/deep learning to foster active and distributed collaboration and ease large-scale image exploitation.
The software can be used remotely, using a web browser, e.g. by life scientists to help them better evaluate drug treatments or understand biological processes using various imaging modalities (including whole-slide tissue images), by pathologists to share and ease their diagnosis (digital pathology), by teachers and students for image-based training purposes (e.g. histology courses), and by computer and data scientists willing to integrate and apply their image recognition algorithms on large imaging datasets using the App Engine or the REST/Python APIs.
In our group, our main contributions are related to (see our latest R&D overview slides):
- Development of novel algorithms, efficient data structures, back-end modules, and web user interfaces for multimodal imaging data
- Development of efficient workflows and novel algorithms (deep learning and tree-based machine learning) for content-based image retrieval, object detection, recognition, and segmentation in very large (multimodal) images
- Reproducible benchmarking of algorithms on realistic datasets
- Applications in biomedical domain with a specific focus on digital pathology and multimodal microscopy datasets
- Applications in any other domain with large sets of large images (geology, digital collections, astronomy, industrial control, …).
- Development of novel algorithms for user behavior analytics
- Software architecture, distributed software deployment, reproducibility, open science, …
In addition to our open-access scientific publications, our results are distributed through the Cytomine open-source software repository.
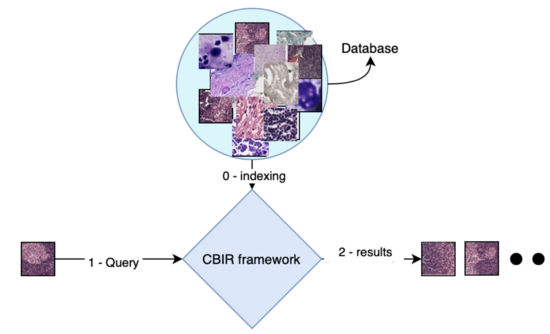
Content-based image retrieval
Algorithms for large-scale, interactive, explainable content-based image retrieval.

Reproducibility and interoperability
Web framework and container-based execution architecture for reproducible deployment and benchmarking of image analysis workflows (Rubens et al., Cell Patterns, 2020).
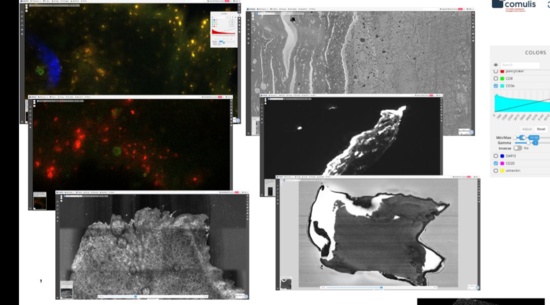
Algorithms and web user interfaces for multimodal datasets
User interfaces and data models for multiple modalities beyond histology incl. MALDI-IMS/multispectral imaging (Rubens et al., Proteomics Clin Appl, 2019).
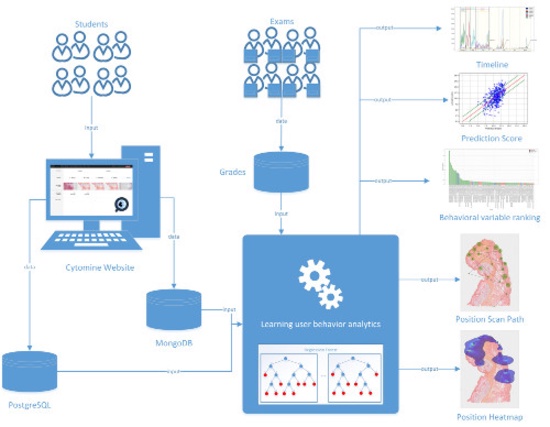
Tools for user behavior analytics
Data analysis modules to study user behavior in educational settings (Fries et al., Anatomical Sciences Education 2023; Vanhee et al., J. Pathology Informatics 2019).
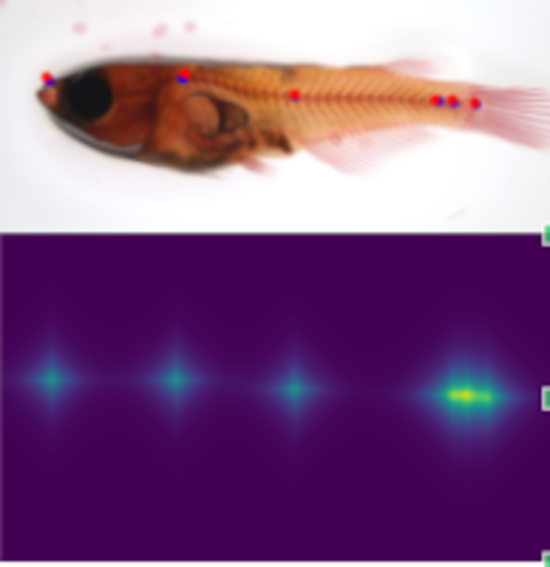
Anatomical landmark detection algorithms
Machine learning algorithms for morphometric change measurements e.g. in developmental and toxicological studies. See e.g. Kumar et al. (Biomolecules 2023, ECCV 2022 Bioimage Computing) and Vandaele et al. (Nature Scientific Reports, 2018).
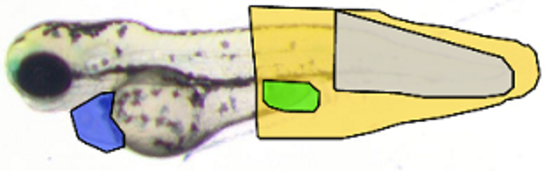
Image classification and object recognition algorithms
Deep/Machine learning algorithms for diagnostic or for phenotyping, e.g. in developmental and toxicological studies. See e.g. Marée et al. (PRL 2016; ISBI 2016); Jeanray et al. (PLOS One 2015); Mormont et al. (CVPRW 2018, IEEE JBHI 2020).
Cell Counting algorithms
Benchmarking of algorithms for cell counting in specific regions of interests within tissues.

Workflows for sorting various types of cells
Image analysis workflows e.g. to detect abnormal cells for early cytological diagnosis. See e.g. Delga et al., Acta Cytologica 2014; Mormont et al., 2016.
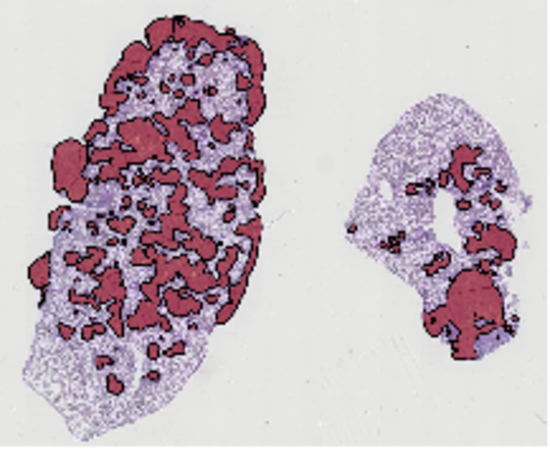
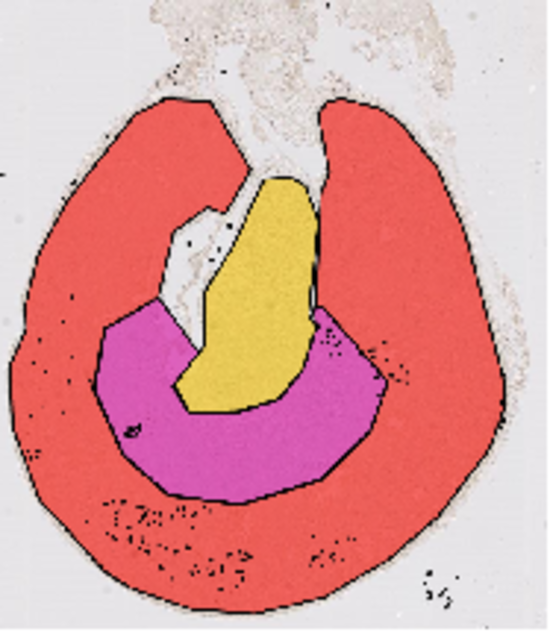
Image segmentation techniques
Self-training algorithms for exploiting sparse annotations (Mormont et al., ECCV AIMIA 2022) and algorithms for the quantification and delineation of tissue areas in whole tissue slides (Marée et al. ISBI 2014).
Latest News
Following Cytomine SA bankruptcy mid-2024, our ULiège team continues the development of the free, open-source (Apache 2.0) Cytomine software through various ongoing research project fundings including EU IMI BigPicture (2021-2027) and Interreg DigiPathConnect (2024-2027). ULiège is also launching a community partnership model for institutions willing to support the project. Cytomine 2025 includes, among other things, a brand new App Engine to run dockerized AI models, a new deployment methodology using Docker Compose (for local deployment) or Kubernetes, and various optimizations since the previous Cytomine Community Edition 2023 release.
Today is the kick-off meeting of the Interreg DigiPathConnect project to foster Cross-border collaboration for digital pathology and AI development. Our team will collaborate with teams from Hasselt, Maastricht, and Köln on software modules for data mining of digital pathology and mass spectrometry imaging data.
Since 2021, we are involved in the BigPicture EU IMI project (2021-2027) to establish a large database of millions of pathology images to accelerate the development of artificial intelligence in medicine. We extend Cytomine web software to be used as the main user interface to visualize and annotate images from the BigPicture repository.
Featured Publication
We kindly ask you to cite this paper when you use Cytomine.
Publications (Methods)
by our team
Publications (Applications)
by collaborators or other teams using Cytomine
Software
(Installation, source code, documentation)
Cytomine can be installed on high-end servers for large-scale studies but also on laptops for small-scale works (then loosing collaboration functionalities).
We maintain the Cytomine repository where you can find installation instructions and documentation.
Access to a demonstration server can be obtained by contacting us.
We kindly ask you to cite Cytomine website url (www.cytomine.org) and our main publication (Marée et al., Bioinformatics 2016) when you use our software in your own work.
Funding
We are/were involved in these projects

BigPicture (2021-2027)
Funded by H2020 Innovative Medicine Initiatives.
This project will establish the biggest database of pathology images to accelerate the development of artificial intelligence in medicine.

DigiPathConnect (2024-2027)
Funded by Interreg Meuse-Rhine Co-funded by the European Union
Fostering Cross-border collaboration for digital pathology and AI development

ARIAC/TRAIL (2021-2026)
Funded by Wallonia/DGO6
ARIAC by DigitalWallonia4.ai / SPW-Recherche. Applications et Recherche pour une Intelligence Artificielle de Confiance

BioMedaqu (2018-2022)
Funded by H2020 MSCA ITN network.
This project aims at developing new image analysis modules for Zebrafish imaging.

COMULIS (2018-2022)
Funded by EU COST Action.
This project aims at developing new multimodal imaging visualization and annotation modules.

IDEES (2017-2020)
Funded by FEDER (ERDF/Wallonia).
This project aimed at developing new big data software modules and deployment mechanisms.

DeepSport (2017-2020)
Funded by Wallonia/DGO6
This project aims at developing new visualization and annotation modules for video data.

NEUBIAS (2016-2020)
Funded by EU COST Action.
This project aimed at developing image analysis reproducible benchmarking modules, see BIAFLOWS.

ADRIC (2017-2021)
Funded by Pole Mecatech, Wallonia/DGO6.
This project aims at developing new AI modules for industrial quality control.

HistoWeb (2014-2017)
Funded by Wallonia/DGO6.
This project aimed at developing Cytomine education software modules

SMASH (2012-2014)
Funded by Wallonia/DGO6.
Cytomine Business development project.

Cytomine (2010-2016)
Funded by Wallonia/DGO6.
Initial Cytomine R&D project.
Contact and Community
We recommend to use the Image.sc forum as the discussion channel for user related questions.
For bugs or feature requests, we recommend to post your issues on Github.
For any questions related to our research activities or for potential research collaborations or internships @ ULiège, feel free to contact our ULiège R&D team:
- uliege@cytomine.org
- +32 4 366 26 44
- Montefiore Institute (B28)
Quartier Polytech 1
Allée de la découverte 10
4000 Liège
Belgium - Office I.127






